Biophysics in Dresden
Research Groups of Biophyics in Dresden

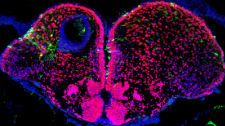 We are interested in understanding how biophysical mechanisms constrain development in the forming vertebrate nervous system. We study morphogen signaling using single molecule methods and live imaging in the living zebrafish embryo, and our wish is to understand how fundamental laws of physics constrain the dynamics of morphogen action and the resulting patterning processes.
We are interested in understanding how biophysical mechanisms constrain development in the forming vertebrate nervous system. We study morphogen signaling using single molecule methods and live imaging in the living zebrafish embryo, and our wish is to understand how fundamental laws of physics constrain the dynamics of morphogen action and the resulting patterning processes.

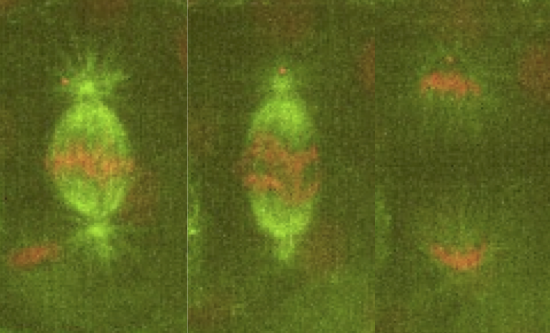 Our lab is interested in studying the underlying principles of self-organization in biological structures. We currently use the metaphase spindle—the protein machinery responsible for segregating the chromosomes into the daughter cells during cell division—from Xenopus laevis egg extract and the zebrafish embryo as model systems to understand the architecture, force generation and scaling of spindles; and chromatin organization in the interphase nucleus. To dissect these processes we combine theory, cell biology, custom build microscopes and biophysical perturbations such as laser ablation.
Our lab is interested in studying the underlying principles of self-organization in biological structures. We currently use the metaphase spindle—the protein machinery responsible for segregating the chromosomes into the daughter cells during cell division—from Xenopus laevis egg extract and the zebrafish embryo as model systems to understand the architecture, force generation and scaling of spindles; and chromatin organization in the interphase nucleus. To dissect these processes we combine theory, cell biology, custom build microscopes and biophysical perturbations such as laser ablation.

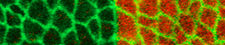 The general interest of our group is to understand the mechanisms by which cells collectively organize to form complex patterns and morphologies in developing tissues. How does collective behavior emerge in cell assemblies? How do mechanical processes like cell adhesion or force generation influence tissue organization? And how are these mechanical processes linked to the chemical signals that orchestrate tissue development? We address these questions by combining genetics with live imaging, quantitative image analysis and biophysical approaches using the fruit fly <i>Drosophila melanogaster</i> as a model organism. Moreover, in collaborations with physicist and mathematicians, we use our quantitative data to build computational models of tissue organization.
The general interest of our group is to understand the mechanisms by which cells collectively organize to form complex patterns and morphologies in developing tissues. How does collective behavior emerge in cell assemblies? How do mechanical processes like cell adhesion or force generation influence tissue organization? And how are these mechanical processes linked to the chemical signals that orchestrate tissue development? We address these questions by combining genetics with live imaging, quantitative image analysis and biophysical approaches using the fruit fly <i>Drosophila melanogaster</i> as a model organism. Moreover, in collaborations with physicist and mathematicians, we use our quantitative data to build computational models of tissue organization.

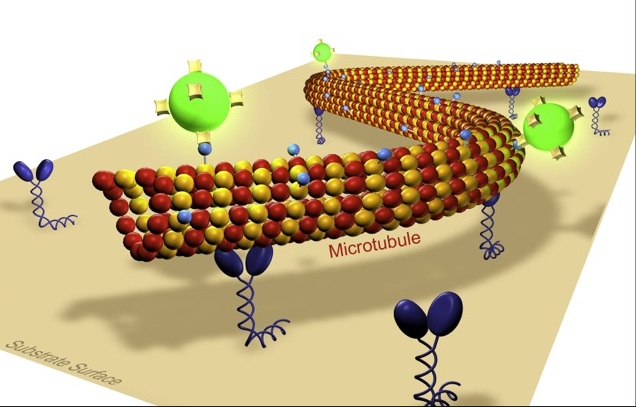 Our lab is interested in the development and the application of novel optical techniques to investigate molecular transport in cell biology and nanotechnology. Building on our experience in single molecule biophysics and in the in vitro reconstruction of subcellular mechano-systems we study cooperative effects in motor transport and cell motility. Moreover we aim to apply biomolecular motor systems in a synthetic, engineered environment for the generation and manipulation of nanostructures. Thereby, our main emphasis is on the development of methods to control the nano-transport sytems by external signals in a spatio-temporal manner. Towards this end we investigate novel biotemplate-based nanostructuring techniques and fabricate smart composite surfaces, where active enzymes are embedded in stimuli-responsive polymer layers.
Our lab is interested in the development and the application of novel optical techniques to investigate molecular transport in cell biology and nanotechnology. Building on our experience in single molecule biophysics and in the in vitro reconstruction of subcellular mechano-systems we study cooperative effects in motor transport and cell motility. Moreover we aim to apply biomolecular motor systems in a synthetic, engineered environment for the generation and manipulation of nanostructures. Thereby, our main emphasis is on the development of methods to control the nano-transport sytems by external signals in a spatio-temporal manner. Towards this end we investigate novel biotemplate-based nanostructuring techniques and fabricate smart composite surfaces, where active enzymes are embedded in stimuli-responsive polymer layers.

 Our group studies how animal tissues grow into their correct morphology during development and how growth can go awry in cancer. We focus on the level of cells-to-tissues, watching how cells move around as they shape a growing tissue and discovering the physical principles underlying their self-organized collective motion. We use Drosophila as a model to decipher the multi-scale regulation of growth during development and human stem cells and cancer organoids to gain insight into the biophysical mechanisms underlying disease.
Our group studies how animal tissues grow into their correct morphology during development and how growth can go awry in cancer. We focus on the level of cells-to-tissues, watching how cells move around as they shape a growing tissue and discovering the physical principles underlying their self-organized collective motion. We use Drosophila as a model to decipher the multi-scale regulation of growth during development and human stem cells and cancer organoids to gain insight into the biophysical mechanisms underlying disease.

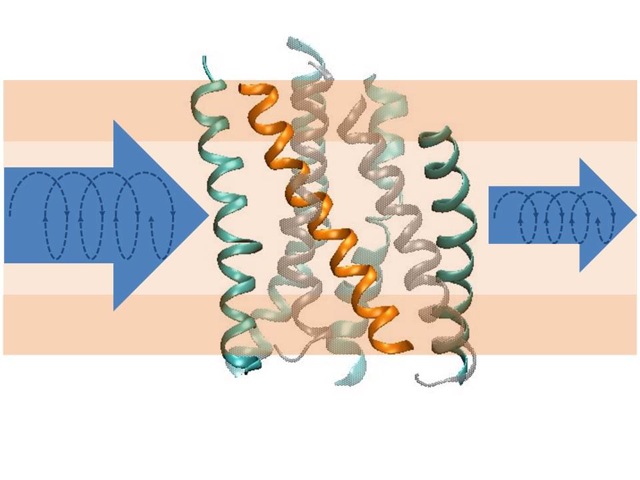 Structural transitions of biomolecules underlie an overwhelming variety of cellular functions. They allow integrating chemically and topologically diverse processes into a regulated network of physico-chemical interactions between macromolecules. Whereas X-ray diffraction provides a snapshot of a static macromolecular structure under crystalline condition, we aim at observing molecular switching processes under native-like conditions and in real time by spectroscopic methods. In combination with CD-spectroscopy and calorimetry, we use infrared spectroscopy as a label-free technique to achieve atomic resolution of conformational transitions on a millisecond to second time scale to address questions that cannot be answered by crystallography, such as the role of dynamic lipid protein interactions in signaling by G-protein coupled receptors or in ion translocation by metal-transporting ATPases. The dynamics of the hydration shell reorganization in these processes is a fundamental physical process that has attracted our attention as it contributes to both structure and energetics of proteins in the complex environment of a biological phase boundary. It has led us to develop novel infrared and fluorescence-based techniques for dynamic analyses of H-bond networks to reach at structurally and energetically consistent descriptions of biomolecular switching and metal binding events in proteins and DNA.
Structural transitions of biomolecules underlie an overwhelming variety of cellular functions. They allow integrating chemically and topologically diverse processes into a regulated network of physico-chemical interactions between macromolecules. Whereas X-ray diffraction provides a snapshot of a static macromolecular structure under crystalline condition, we aim at observing molecular switching processes under native-like conditions and in real time by spectroscopic methods. In combination with CD-spectroscopy and calorimetry, we use infrared spectroscopy as a label-free technique to achieve atomic resolution of conformational transitions on a millisecond to second time scale to address questions that cannot be answered by crystallography, such as the role of dynamic lipid protein interactions in signaling by G-protein coupled receptors or in ion translocation by metal-transporting ATPases. The dynamics of the hydration shell reorganization in these processes is a fundamental physical process that has attracted our attention as it contributes to both structure and energetics of proteins in the complex environment of a biological phase boundary. It has led us to develop novel infrared and fluorescence-based techniques for dynamic analyses of H-bond networks to reach at structurally and energetically consistent descriptions of biomolecular switching and metal binding events in proteins and DNA.

 Many physiological processes rely on a well-orchestrated sequence of cellular shape changes. Important examples include cell division, cell migration and the morphogenesis of tissues. To obtain a mechanistic understanding of such cellular shape changes, we strive to characterize cells and tissues as an active material with certain macroscopic mechanical properties. In particular, our group works on the development of theoretical and experimental tools for the quantification of active material properties of cells and tissues. Furthermore, we investigate how these active material properties emerge from underlying molecular constituents inside cells and inside the extracellular material.
Many physiological processes rely on a well-orchestrated sequence of cellular shape changes. Important examples include cell division, cell migration and the morphogenesis of tissues. To obtain a mechanistic understanding of such cellular shape changes, we strive to characterize cells and tissues as an active material with certain macroscopic mechanical properties. In particular, our group works on the development of theoretical and experimental tools for the quantification of active material properties of cells and tissues. Furthermore, we investigate how these active material properties emerge from underlying molecular constituents inside cells and inside the extracellular material.Current research foci in our group are mitotic rounding, epithelial-mesenchymal transition, force-sensing actin cross-linkers, active gels and epithelial folding.

 Our theory group wants to understand how cells and tissues robustly process information and form functional structures, using tools from nonlinear dynamics, statistical physics and information theory in close collaborations with experimentalists. Biological systems of interest include the dynamics of cilia, cellular navigation and self-organized pattern formation during embryonic development and regeneration – e.g. how muscle cells form crystal-like myofibrils, diatom cells build their intricate glass shells, or axolotl regrow lost limbs of correct size.
Our theory group wants to understand how cells and tissues robustly process information and form functional structures, using tools from nonlinear dynamics, statistical physics and information theory in close collaborations with experimentalists. Biological systems of interest include the dynamics of cilia, cellular navigation and self-organized pattern formation during embryonic development and regeneration – e.g. how muscle cells form crystal-like myofibrils, diatom cells build their intricate glass shells, or axolotl regrow lost limbs of correct size.

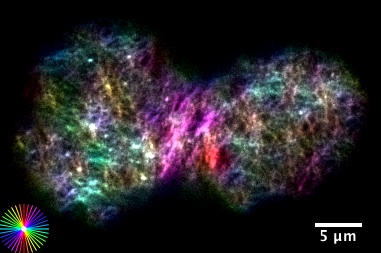 Morphogenesis refers to the generation of form in Biology. We are interested in bridging physical mechanisms from molecular scales to cell and tissue scales, to understand how an unpatterned blob of cells develops into a fully structured and formed organism. We combine theory and experiment, and investigate force generation on multiple scales. At the level of cells an tissues we study how the actomyosin cell cortex self contracts, reshapes and deforms, and how these morphogenetic activities couple to regulatory biochemical pathways. At the level of molecules we investigate force generation and movement of individual molecules of RNA polymerases in the context of gene expression and transcriptional proofreading
Morphogenesis refers to the generation of form in Biology. We are interested in bridging physical mechanisms from molecular scales to cell and tissue scales, to understand how an unpatterned blob of cells develops into a fully structured and formed organism. We combine theory and experiment, and investigate force generation on multiple scales. At the level of cells an tissues we study how the actomyosin cell cortex self contracts, reshapes and deforms, and how these morphogenetic activities couple to regulatory biochemical pathways. At the level of molecules we investigate force generation and movement of individual molecules of RNA polymerases in the context of gene expression and transcriptional proofreading

 Our aim is to understand how epithelial tissue forms de novo. The transition of progenitor stem cells into a polarized epithelium is a fundamental process during embryogenesis and the formation of many organs. Our approach is to use 3D cell culture organoids as model systems to dissect the underlying mechanisms of cell polarization and differentiation during this transition. The long-term goal is to integrate cell adhesion, cortical flows and membrane trafficking into a model which describes the transition process as a self-organizing system. To characterize the transition from cells to tissue we use optical microscopy (from STED to SPIM) in combination with genetic engineering. We complement this by reconstitutions of self-organization on artificial membranes and controlled cell adhesion assays.
Our aim is to understand how epithelial tissue forms de novo. The transition of progenitor stem cells into a polarized epithelium is a fundamental process during embryogenesis and the formation of many organs. Our approach is to use 3D cell culture organoids as model systems to dissect the underlying mechanisms of cell polarization and differentiation during this transition. The long-term goal is to integrate cell adhesion, cortical flows and membrane trafficking into a model which describes the transition process as a self-organizing system. To characterize the transition from cells to tissue we use optical microscopy (from STED to SPIM) in combination with genetic engineering. We complement this by reconstitutions of self-organization on artificial membranes and controlled cell adhesion assays.

 We are interested in studying cell polarity, centrosome assembly and microtubule dynamics. Currently, the main goal of the lab is to understand how cells form non-membrane bound compartments. Using a combination of genetics and physics, we have discovered that a principle underlying the organization of many of these compartments is liquid-liquid phase separation. We study these compartments both in vivo and in vitro, using reconstitution methods. We primarily use C. elegans as a model system, but we complement this with studies in human iPS cells.
We are interested in studying cell polarity, centrosome assembly and microtubule dynamics. Currently, the main goal of the lab is to understand how cells form non-membrane bound compartments. Using a combination of genetics and physics, we have discovered that a principle underlying the organization of many of these compartments is liquid-liquid phase separation. We study these compartments both in vivo and in vitro, using reconstitution methods. We primarily use C. elegans as a model system, but we complement this with studies in human iPS cells.

 Biological Physics at PKS focuses on the theoretical study of active processes in cells and
Biological Physics at PKS focuses on the theoretical study of active processes in cells andtissues. We develop concepts and methods to understand principles that govern the organization
of cellular processes and the morphogenesis of tissues. To this end approaches from statistical
physics of non-equilibrium systems and from non-linear dynamics are very important. Key to our
work are close collaborations with experimental groups, in particular, at the Max Planck Institute
of Molecular Cell Biology and Genetics, Dresden.

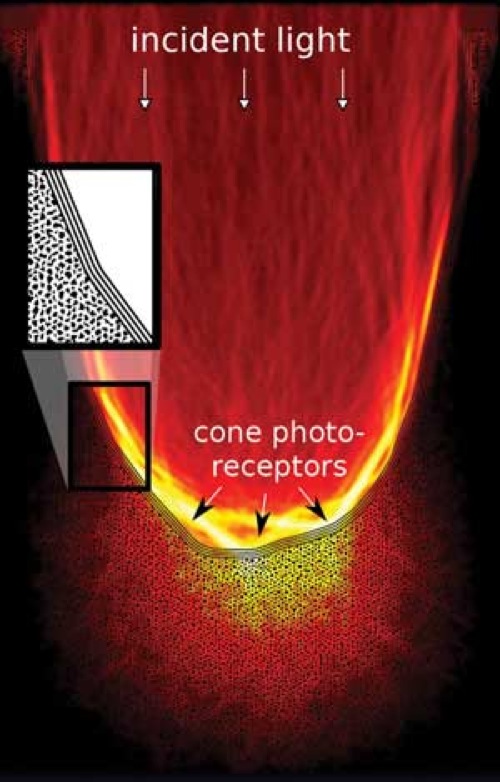 Our research addresses one of the retinas most surprising, but least investigated characteristics, its optical architecture: since the sensitive portions of the photoreceptor cells are found on the back of the vertebrate retina, light needs to travel through several layers of living neuronal tissue prior to detection. What is usually regarded as being a problem of neuronal activity is complemented from the perspective of optics, focusing on one key question: how does the retina deal with incident light?
Our research addresses one of the retinas most surprising, but least investigated characteristics, its optical architecture: since the sensitive portions of the photoreceptor cells are found on the back of the vertebrate retina, light needs to travel through several layers of living neuronal tissue prior to detection. What is usually regarded as being a problem of neuronal activity is complemented from the perspective of optics, focusing on one key question: how does the retina deal with incident light?For this we are using custom design microscopy to gain a detailed understanding of how optical constrains shape retinal development, from the overall architecture down to the level of the chromatin organization. Apart from its importance for the initiation of the visual process, light propagation in neuronal tissues is also key to the optical observation of brain activity over large scales. The experimental side of this research is accompanied by theoretical approaches and computer modeling.
Further interests of our lab address reaction-diffusion systems of pattern formation, and the origin of life.
 Evolution brings us incredible diversity in animal size and shape, but remarkably, our organs and limbs remain proportional to our body size. This entails an extraordinary level of coordination across different scales. How do organs measure and control their size? This is a crucial question that has remained long unresolved in Biology. We aim to answer it by looking at cells. We are focused on studying biophysical modes of cellular communication: Electrical Flows, Chemical Signalling and Mechanical Forces, with the goal of understanding how organ growth information is encoded.
Evolution brings us incredible diversity in animal size and shape, but remarkably, our organs and limbs remain proportional to our body size. This entails an extraordinary level of coordination across different scales. How do organs measure and control their size? This is a crucial question that has remained long unresolved in Biology. We aim to answer it by looking at cells. We are focused on studying biophysical modes of cellular communication: Electrical Flows, Chemical Signalling and Mechanical Forces, with the goal of understanding how organ growth information is encoded. We use the zebrafish larva as an in vivo model, as it allows for optimal quantitative live imaging and amenable genetics. Importantly, zebrafish regenerate their organs! This allows us to search for common rules of growth: not only in Development, but also in Regeneration. Because of this, we can choose to perform our experiments with Fast or Slow growth rates – at Steady state or Out of Equilibrium scenarios – achieving proportionality or recovering it! We generate the molecular tools that enable us to measure and manipulate the physical parameters at play during organ growth. We also like to collaborate closely with theoreticians as well as computer scientists, so that together, we can get further mechanistic insight into how do organs grow.

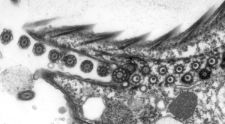 Cryo-transmission electron microscopy (cryo-EM) is a powerful technology that can be used to reveal the three-dimensional architecture and the assembly of macromolecular machines, organelles and cells. We use cryo-EM and electron tomography to reveal the structural basis of flagella and cilia assembly, as well as motility. The assembly takes place at the distal tip of the cilium and the transport of ciliary proteins from the cell body to the tip is mediated by the intraflagellar transport (IFT) machinery. We currently study the assembly of the cilium by investigating the detailed 3D structure, the molecular arrangement, the protein composition, and the dynamics of the ciliary tip complex and of the IFT trains.
Cryo-transmission electron microscopy (cryo-EM) is a powerful technology that can be used to reveal the three-dimensional architecture and the assembly of macromolecular machines, organelles and cells. We use cryo-EM and electron tomography to reveal the structural basis of flagella and cilia assembly, as well as motility. The assembly takes place at the distal tip of the cilium and the transport of ciliary proteins from the cell body to the tip is mediated by the intraflagellar transport (IFT) machinery. We currently study the assembly of the cilium by investigating the detailed 3D structure, the molecular arrangement, the protein composition, and the dynamics of the ciliary tip complex and of the IFT trains.

 We combine expertise from computer science, mathematics, physics and biology in order to develop and apply computational methods for the study of spatiotemporal biological processes in 3D. We exploit the unifying framework of particle methods for numerical simulation, image analysis, and model identification. Since computational biology comes with a unique set of challenges, from complex geometric shapes to non-equilibrium processes, we develop and apply novel computational methods in a targeted co-design approach with the ultimate mission of understanding the algorithms of tissue formation.
We combine expertise from computer science, mathematics, physics and biology in order to develop and apply computational methods for the study of spatiotemporal biological processes in 3D. We exploit the unifying framework of particle methods for numerical simulation, image analysis, and model identification. Since computational biology comes with a unique set of challenges, from complex geometric shapes to non-equilibrium processes, we develop and apply novel computational methods in a targeted co-design approach with the ultimate mission of understanding the algorithms of tissue formation.

 Our group is interested in the physical mechanisms that underlie the organization of the genetic material. We are currently investigating how the sequence-dependent mechanics and geometry of the DNA double helix affects its compaction into DNA-protein complexes known as nucleosomes. In addition, we want to understand how the positioning along the DNA of nucleosomes and their dynamics are influenced by active motor proteins called chromatin remodelers. On a larger scale, we study the organization and dynamics of DNA in terms of polymer physics combined with the phenomena of loop extrusion and macromolecular condensates.?
Our group is interested in the physical mechanisms that underlie the organization of the genetic material. We are currently investigating how the sequence-dependent mechanics and geometry of the DNA double helix affects its compaction into DNA-protein complexes known as nucleosomes. In addition, we want to understand how the positioning along the DNA of nucleosomes and their dynamics are influenced by active motor proteins called chromatin remodelers. On a larger scale, we study the organization and dynamics of DNA in terms of polymer physics combined with the phenomena of loop extrusion and macromolecular condensates.?

 Our lab is curious about nature and its huge pool of molecular machines. We are studying these <i>in vitro</i> to understand their chemo-mechanical and enzymatic activities using fluorescence- and force-based single-molecule techniques. In particular, we are interested in DNA interacting enzymes that are involved in DNA replication and recombination. Furthermore, we are studying how cytosolic and membrane proteins find their tertiary structure and loose it in the process of protein degradation. To this end, we are developing novel tools and techniques based on optical and mechanical manipulation combined with fluorescence based single-molecule FRET studies.
Our lab is curious about nature and its huge pool of molecular machines. We are studying these <i>in vitro</i> to understand their chemo-mechanical and enzymatic activities using fluorescence- and force-based single-molecule techniques. In particular, we are interested in DNA interacting enzymes that are involved in DNA replication and recombination. Furthermore, we are studying how cytosolic and membrane proteins find their tertiary structure and loose it in the process of protein degradation. To this end, we are developing novel tools and techniques based on optical and mechanical manipulation combined with fluorescence based single-molecule FRET studies.

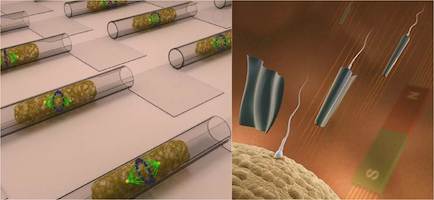 We explore the possibilities of micro- and nanotechnologies towards some of their most fascinating biomedical application scenarios. For instance, we create ultra-compact microfluidic systems for single cell manipulation and analysis as well as flexible and soft electronic systems for a new generation of implant materials. We assemble and operate biomedical microrobots, which are remotely controlled and envisioned to serve for non-invasive microsurgical tasks and targeted drug delivery. The combination of a biological power source (e.g. a spermatozoon) and a microdevice (e.g. a magnetic microtube) is a compelling approach for fascinating future applications such as assisted in-vivo artificial fertilization and gynecological cancer screening and treatment.
We explore the possibilities of micro- and nanotechnologies towards some of their most fascinating biomedical application scenarios. For instance, we create ultra-compact microfluidic systems for single cell manipulation and analysis as well as flexible and soft electronic systems for a new generation of implant materials. We assemble and operate biomedical microrobots, which are remotely controlled and envisioned to serve for non-invasive microsurgical tasks and targeted drug delivery. The combination of a biological power source (e.g. a spermatozoon) and a microdevice (e.g. a magnetic microtube) is a compelling approach for fascinating future applications such as assisted in-vivo artificial fertilization and gynecological cancer screening and treatment.

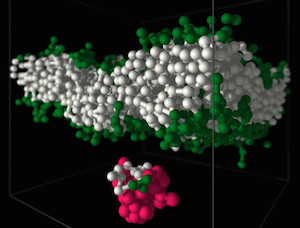 Polymers are most important molecules in living systems and an essential component of materials ranging from packing to smart surfaces and applications of synthetic polymers in contact with living matter. Our group at the Leibniz-Institute of Polymer Research Dresden (IPF) is studying the physical properties of polymers using theoretical concepts and computer simulations. The long-chain nature, flexibility, architectural diversity and the multitude of possible forms of interactions and self-organization in polymer systems is a challenge for analytical approaches and demands for the development of new computational algorithms. Very important for our research is the close collaboration with experimental groups. Our research interests in the field of bio-functional polymers concerns in particular the interactions of polymers with lipid membranes and bio-functional polymer gels.
Polymers are most important molecules in living systems and an essential component of materials ranging from packing to smart surfaces and applications of synthetic polymers in contact with living matter. Our group at the Leibniz-Institute of Polymer Research Dresden (IPF) is studying the physical properties of polymers using theoretical concepts and computer simulations. The long-chain nature, flexibility, architectural diversity and the multitude of possible forms of interactions and self-organization in polymer systems is a challenge for analytical approaches and demands for the development of new computational algorithms. Very important for our research is the close collaboration with experimental groups. Our research interests in the field of bio-functional polymers concerns in particular the interactions of polymers with lipid membranes and bio-functional polymer gels.

 Biology is well equipped in exploiting a large number of out of equilibrium processes to support life. A complete understanding of these mechanisms is still in its infancy due to the complexity and number of the individual components involved in the reactions. However, a bottom up approach allows us to replicate key biological processes using a small number of basic building blocks. This methodology has the added advantage that properties and characteristics of the artificial cell can be readily tuned and adapted. Working between biophysics, materials science and synthetic biology, we reimagine and translate the physical phenomena which drive out of equilibrium processes in cells into novel, robust and dynamic systems for synthetic biology applications.
Biology is well equipped in exploiting a large number of out of equilibrium processes to support life. A complete understanding of these mechanisms is still in its infancy due to the complexity and number of the individual components involved in the reactions. However, a bottom up approach allows us to replicate key biological processes using a small number of basic building blocks. This methodology has the added advantage that properties and characteristics of the artificial cell can be readily tuned and adapted. Working between biophysics, materials science and synthetic biology, we reimagine and translate the physical phenomena which drive out of equilibrium processes in cells into novel, robust and dynamic systems for synthetic biology applications.

 We are interested in the collective dynamics and self-organization of distributed systems exhibiting multiple feedback. We combine and develop theoretical concepts from Nonlinear Dynamics, Graph Theory and Statistical Physics, Applied Mathematics and computational tools to understand mechanisms underlying collective phenomena in biological and engineered systems. Our research focus is on the dynamics of networked systems. Application areas include distributed biological computing, structure function relations in biological networks and the dynamics of biological and engineered flow networks.
We are interested in the collective dynamics and self-organization of distributed systems exhibiting multiple feedback. We combine and develop theoretical concepts from Nonlinear Dynamics, Graph Theory and Statistical Physics, Applied Mathematics and computational tools to understand mechanisms underlying collective phenomena in biological and engineered systems. Our research focus is on the dynamics of networked systems. Application areas include distributed biological computing, structure function relations in biological networks and the dynamics of biological and engineered flow networks.

 In order for the cell to function properly, proteins must be robust to both changes in their environment and errors made during their synthesis. At the same time, proteins also need to be able to evolve novel functions to survive on long evolutionary timescales. The very same processes, i.e. genetic and phenotypic mutations, generate the diversity that leads to functional innovations and broken proteins, ultimately resulting in novel organisms, diseases and, in some cases, extinction. We aim to define and quantify the phenotypic plasticity of a protein, and to identify the compensatory mechanisms that buffer otherwise deleterious mutations. We wish to reveal the evolutionary potential of latent phenotypes to create novel functions and to influence gene-disease associations.
In order for the cell to function properly, proteins must be robust to both changes in their environment and errors made during their synthesis. At the same time, proteins also need to be able to evolve novel functions to survive on long evolutionary timescales. The very same processes, i.e. genetic and phenotypic mutations, generate the diversity that leads to functional innovations and broken proteins, ultimately resulting in novel organisms, diseases and, in some cases, extinction. We aim to define and quantify the phenotypic plasticity of a protein, and to identify the compensatory mechanisms that buffer otherwise deleterious mutations. We wish to reveal the evolutionary potential of latent phenotypes to create novel functions and to influence gene-disease associations.